Martin Balastik Group
Institute of Physiology, CAS
TPC group associated to DIGS-ILS

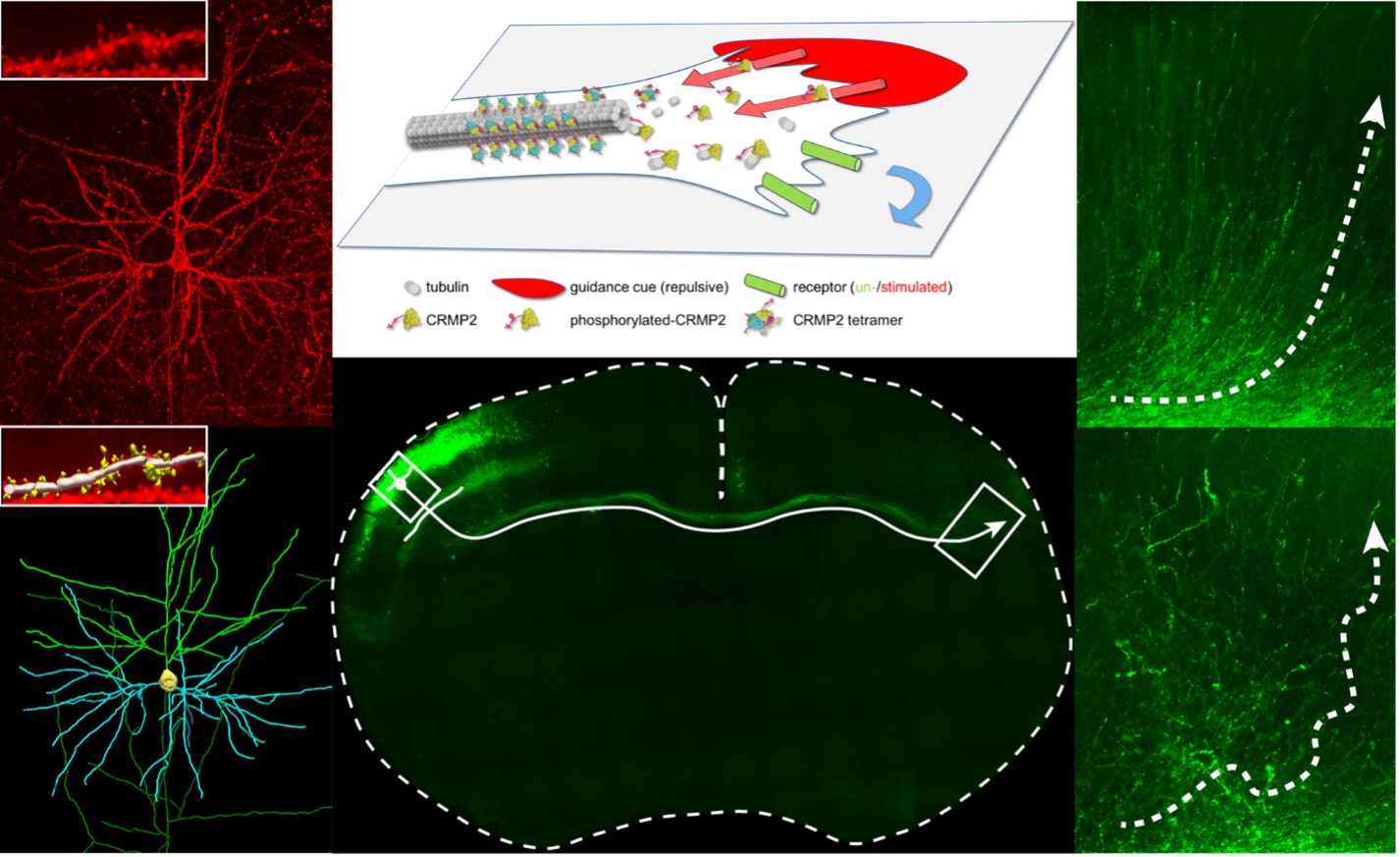
During development of human brain 86 billion neurons form over 1014 connections (synapses). Precision of these connections is critical for the brain function and changes of brain connectivity have been associated with several neurodevelopmental disorders as autism, schizophrenia or epilepsy. Neuron connections are generated through processes of axon/dendrite guidance, synapse formation and refinement. Their growth is controlled at the extracellular level by gradients of attractive or repulsive guidance cues, which are translated into appropriate changes of cytoskeletal dynamics within the growing neurons.
Our laboratory is interested in the regulation of development of the brain connectivity. We characterize the molecular mechanisms responsible for the precise translation of the extrinsic guidance signals into changes of microtubule (MT) dynamics, enabling a fine-tuned response of every growing neuron projection and allowing it to reach its correct cellular target. We focus on the role of microtubule-associated proteins (MAPs) and tubulin post-translational modifications (PTMs) and utilize a combination of in vitro MT systems and primary neuron cultures to characterize the molecular and biological functions of various MAP isoforms and tubulin PTMs, as well as their mutual regulation. Furthermore, we generate and analyze new germinal (knock-out) and somatic (in utero electroporation) mouse mutants and analyze how different splice variants of MAPs in combination with tubulin PTMs affect neuron polarization, migration and cortical development in embryonic development. To characterize their role in adult mice, we use life intravital microscopy of cortical pyramidal neurons through cranial windows. The combination of model systems and techniques provides a detail information about the molecular function and developmental role of MAP isoforms and tubulin PTMs as well as their relevance for pathogenesis of neurodevelopmental disorders.
FUTURE PROJECTS
- MAPs in axon growth, guidance and pruning – an isoform-specific role of CRMP (Collapsin response mediator proteins) protein family.
- Prolyl isomerases in neural development - conformational regulation of CRMPs by prolyl isomerases and its role in neural development.
- Combinatorial effect of tubulin PTMs and MAPs on the regulation of neuron polarization, migration and pruning in vivo/in vitro.
Institute of Physiology, Czech Academy of Sciences
Videnska 1083
14000 Prague 4
Czech Republic
since 2014
Group leader at the Institute of Physiology, Czech Academy of Sciences, Prague, Czech Republic
2012-2013
Principal investigator at the Institute of Molecular Genetics, Prague, Czech Republic
2004 -2011
Postdoc at Harvard Medical School, Beth Israel Deaconess Medical Center, Boston, USA
2003
PhD in Neuroscience, International Max Planck Research School, Göttingen, Germany
Ziak J, Weissova R, Jerabkova K, Janikova M, Maimon R, Petrasek T, Pukajova B, Kleisnerova M, Wang M, Brill MS, Kasparek P, Zhou X, Alvarez-Bolado G, Sedlacek R, Misgeld T, Stuchlik A, Perlson E, Balastik M.
CRMP2 mediates Sema3F-dependent axon pruning and dendritic spine remodeling
EMBO REPORTS. 2020; 21(3); e48512
Magiera MM, Bodakuntla S, Ziak J, Lacomme S, Sousa PM, Leboucher S, Hausrat TJ, Bosc C, Andrieux A, Kneussel M, Landry M, Calas A, Balastik M, Janke C.
Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport.
EMBO Journal 2018, 37(23), e100440
Balastik M, Zhou XZ, Alberich-Jorda M, Weissova R, Ziak J, Pazyra-Murphy MF, Cosker KE, Machonova O, Kozmikova I, Chen CH, Pastorino L, Asara JM, Cole A, Sutherland C, Segal RA, Lu KP.
Prolyl Isomerase Pin1 Regulates Axon Guidance by Stabilizing CRMP2A Selectively in Distal Axons
Cell Reports. 2015, 13(4), 812-828
Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, Shaik S, Lee DY, Finn G, Balastik M, Chen CH, Luo M, Tron AE, Decaprio JA, Zhou XZ, Wei W, Lu KP.
Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase
Mol Cell. 2012 Jun 29;46(6):771-83